What is JASPAR?
JASPAR is a regularly maintained open-access database storing manually curated transcription factors (TF) binding profiles as position frequency matrices (PFMs). PFMs summarize occurrences of each nucleotide at each position in a set of observed TF-DNA interactions. PFMs can be transformed to probabilistic or energistic models to construct position weight matrices (PWMs) or position-specific scoring matrices (PSSMs), which can be used to scan any DNA sequence to predict TF binding sites (TFBSs). The JASPAR database provides TFBSs predicted using the profiles in the CORE collection.
The motifs in JASPAR are collected in two ways:
- Internally: de novo generated motifs, by analyzing ChIP-seq/-exo sequences using a custom motif discovery pipeline (check the code at our repository).
- Externally: motifs taken directly from other publications and/or resources.
In both cases, the selected motifs are manually curated. Specifically, our curators assess the quality of the motif and search for an orthogonal publication providing support to the motif as the bona fide motif recognized by the TF of interest (e.g., a motif found in ChIP-seq peaks looks similar to one found by SELEX-seq). The Pubmed ID associated with the orthogonal support is provided in the TF profile metadata.
JASPAR is the only database with this scope where the data can be used with no restrictions (open source). For a comprehensive review of models and how they can be used, please see the following reviews
JASPAR collections
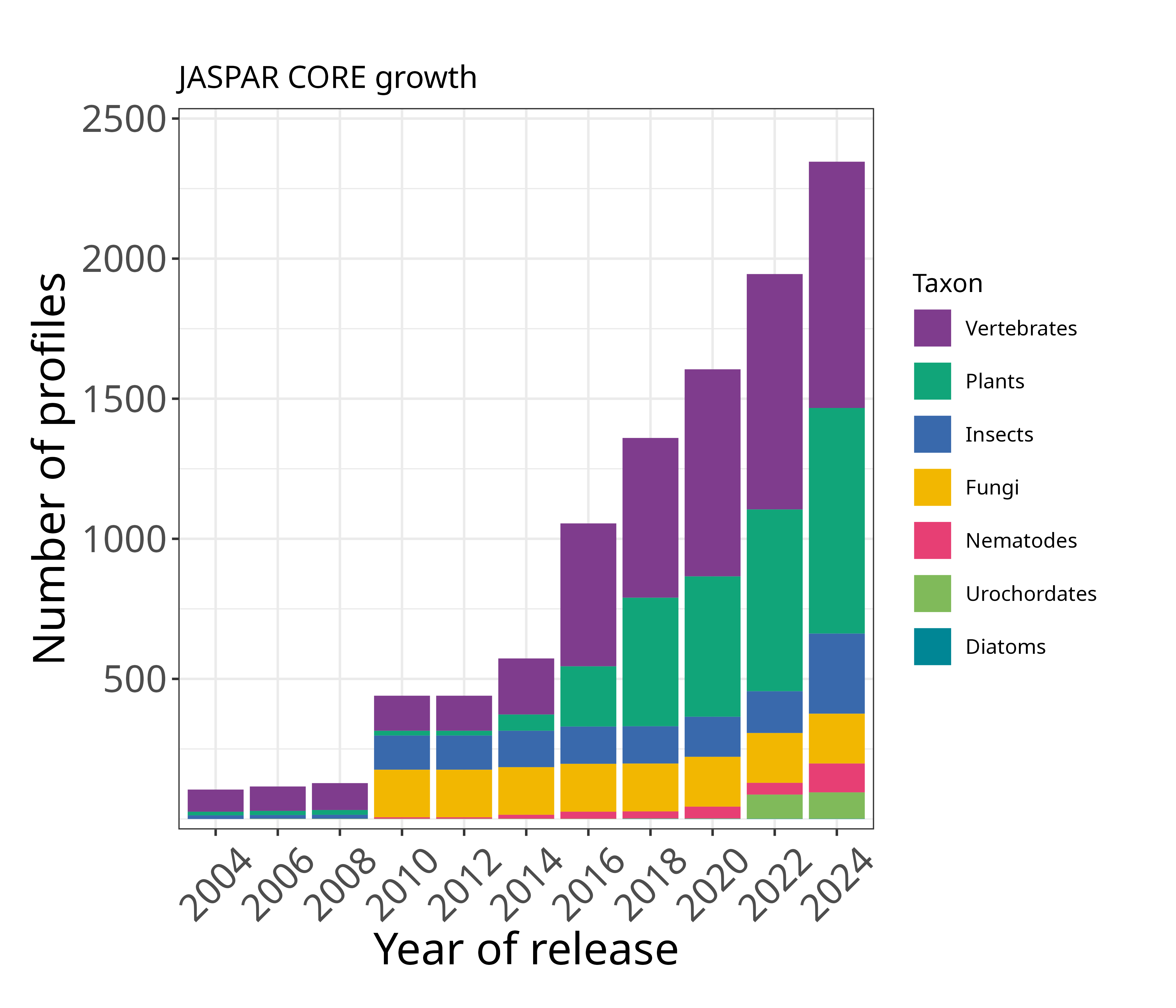
The JASPAR CORE database contains a curated, non-redundant set of profiles, derived from published collections of experimentally defined transcription factor binding sites for eukaryotes. The prime difference to similar resources (TRANSFAC, etc.) consist of the open data access, non-redundancy and quality.
When should it be used? When seeking models for specific factors or structural classes, or if experimental evidence is paramount
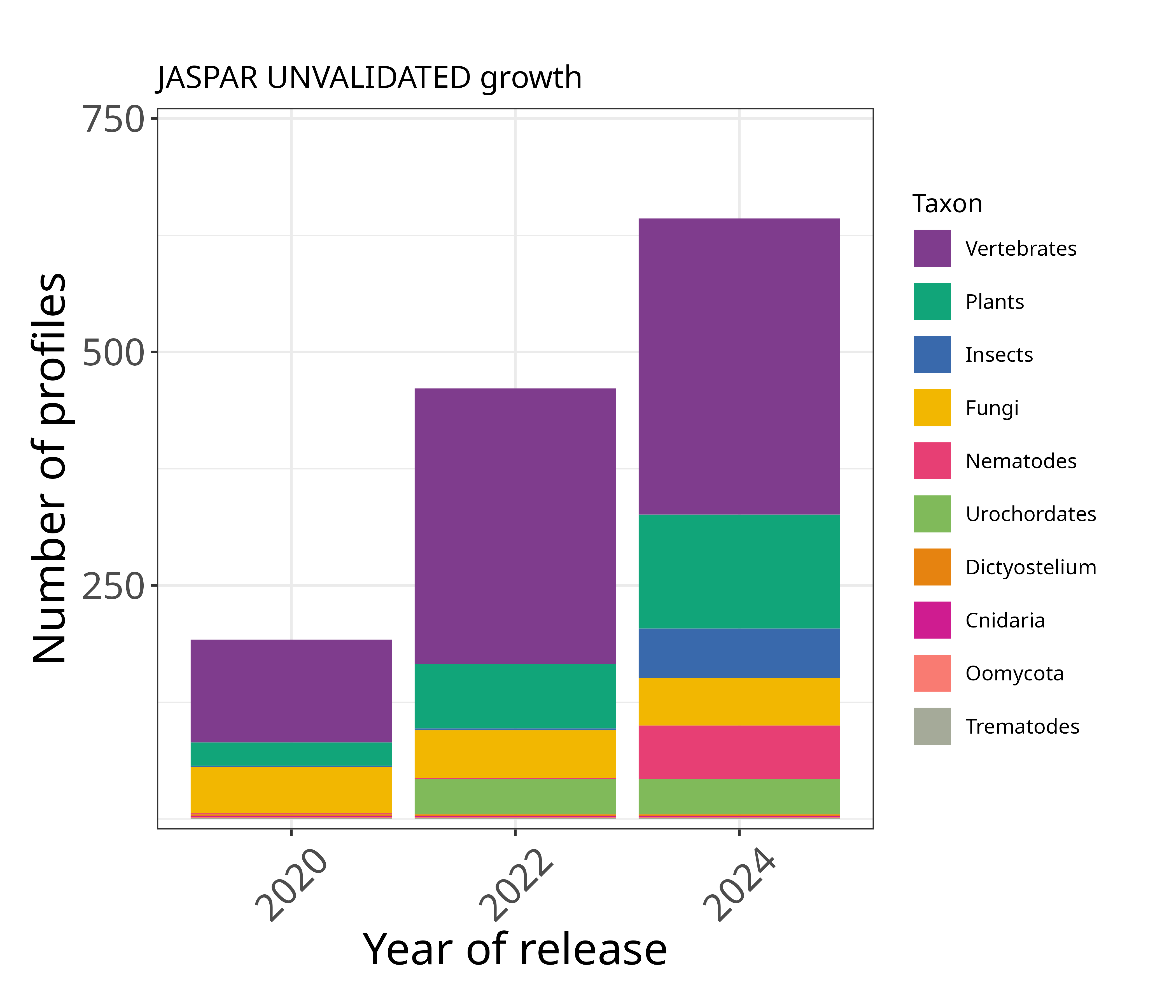
These profiles are regarded as unvalidated because our curators failed to find any orthogonal support from existing literature. We encourage the community to perform experiments and/or point us to literature that our curators missed in order to support these profiles.
When should it be used? These profiles are not non-validated so we recommend not to use them.