Frequently Asked Questions
How do I cite JASPAR?
If you simply want to acknowledge that you used the latest version of the database, use this citation:
Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Chèneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles Nucleic Acids Res. in_press; doi: 10.1093/nar/gkad1059
The complete list of citations are as follows:
- The original JASPAR paper:
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles Nucleic Acids Res. 2004 Jan 1;32(Database issue):D91-4.; doi: 10.1093/nar/gkh012
- The first extension (JASPAR FAM and PHYLOFACTS collections):
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles Nucleic Acids Res. 2006 Jan 1;34(Database issue):D95-7.; doi: 10.1093/nar/gkj115
- Second expansion (POLII, SPLICE, CNE, many changes in the web service including matrix permutations):
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update Nucleic Acids Res. 2008 Jan;36(Database issue):D102-6.; doi: 10.1093/nar/gkm955
- Third expansion (Large expansion of the CORE collection, including yeast and worm matrices. Also includes new PBM collections):
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles Nucleic Acids Res. 2010 Jan;38(Database issue):D105-10.; doi: 10.1093/nar/gkp950
- Fourth expansion:
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen C-y, Chou A, Ienasescu H, Lim J, Shyr C, Tan G, Zhou M, Lenhard B, Sandelin A, Wasserman WW. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles Nucleic Acids Res. 2014 Jan;42(Database issue):D142-7.; doi: 10.1093/nar/gkt997
- Fifth expansion:
- Mathelier A, Fornes O, Arenillas DJ, Chen C, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman W. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles Nucleic Acids Res. 2016 44: D110-D115; doi: 10.1093/nar/gkv1176
- Sixth expansion (new web and REST interface):
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F, Mathelier A. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework Nucleic Acids Res. 2018; 46:D260–D266.; doi: 10.1093/nar/gkx1126
- Seventh expansion (introducing unvalidated TF-binding profiles):
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, Santana-Garcia W, Tan G, Chèneby J, Ballester B, Parcy F, Sandelin A, Lenhard B, Wasserman WW, Mathelier A. JASPAR 2020: update of the open-access database of transcription factor binding profiles Nucleic Acids Res. 2019; doi: 10.1093/nar/gkz1001
- Eighth expansion:
- Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Berhanu Lemma R, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, Fornes O, Leung TY, Aguirre A, Hammal F, Schmelter D, Baranasic D, Ballester B, Sandelin A, Lenhard B, Vandepoele K, Wasserman WW, Parcy F, Mathelier A JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles Nucleic Acids Res. 2022 Jan 7;50(D1):D165-D173.; doi: 10.1093/nar/gkab1113
- Ninth expansion:
- Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Chèneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles Nucleic Acids Res. in_press; doi: 10.1093/nar/gkad1059
What motif formats does JASPAR support?
The data from JASPAR can be downloaded in four different motif formats: raw, JASPAR, TRANSFAC, and MEME. Read more about motif formats in the JASPAR documentation here.
How are TFBSs for a profile in JASPAR computed?
JASPAR provides genomic TFBS predictions for 8 organisms (Arabidopsis thaliana, Caenorhabditis elegans, Ciona intestinalis, Danio rerio, Drosophila melanogaster, Homo sapiens, Mus musculus, and Saccharomyces cerevisiae) with the JASPAR CORE PFMs associated with the same taxon.
DNA sequences were scanned with JASPAR CORE TF-binding profiles for each taxa independently using PWMScan. We selected TFBS predictions with a PWM relative score ≥ 0.8 and a p-value < 0.05.
How should I interpret the TFBSs score?
The score (weight) of each binding site is calculated with the following formula:
Weight = log( P(Site|Matrix) / P(Site|Background) )
Where:
- P(Site|Matrix) corresponds to the probability of observing a sequence given the frequencies at the PFMs.
- P(Site|Background) corresponds to the probability of observing a sequence given the frequencies described by the background model.
The range of weights observed in long matrices (e.g., REST, with 21 nt) will be higher than those observed in shorter matrices (SOX2, 11 nt), for this reason a threshold based directly on the weight is not optimal. Since each motif has its own size and information content, this critically influences the expected distribution of weights.
There are two solutions to overcome this challenge:
- Normalize the weight scores. For example, a threshold of W=5 could be good for SOX2 but will be far from optimal for REST, so, to avoid this problem the weights (or scores) are normalized where the highest weight (score) corresponds to a Relative score of 100 or 1000 (depending the granularity). A Relative score of 800 will consider all the binding sites above that number independently of the matrix width and the weight, maybe a Score of 800 in Sox2 corresponds to a weight of 5, whilst in REST corresponds to a weight of 12.
- Calculate a site P-value. This gives a better intuition about the risk associated with each prediction. See 10.1186/1748-7188-2-15 and 10.1038/nprot.2008.97.
The relative score Srel is computed as Srel = (W - min) / (max - min) where W is the score of the sequence given the PWM, min (max) is the minimal (maximal) score that can be obtained from the PWM.
Where do I download TFBSs for a profile in JASPAR?
The TFBS predictions associated with all PFMs are available here (note that they are not available to download through this website). We provide JASPAR TFBS predictions as genomic tracks, which can be visualized in genome browsers. Notably, the UCSC Genome Browser now presents predicted human JASPAR TFBS data as a native track for the human genome with information such as TF names, TFBS prediction scores, and PFM logo for each of the 12+ billion predictions. For details check documentation for genomic tracks.
Where do I download the sequences used to construct the JASPAR PFMs?
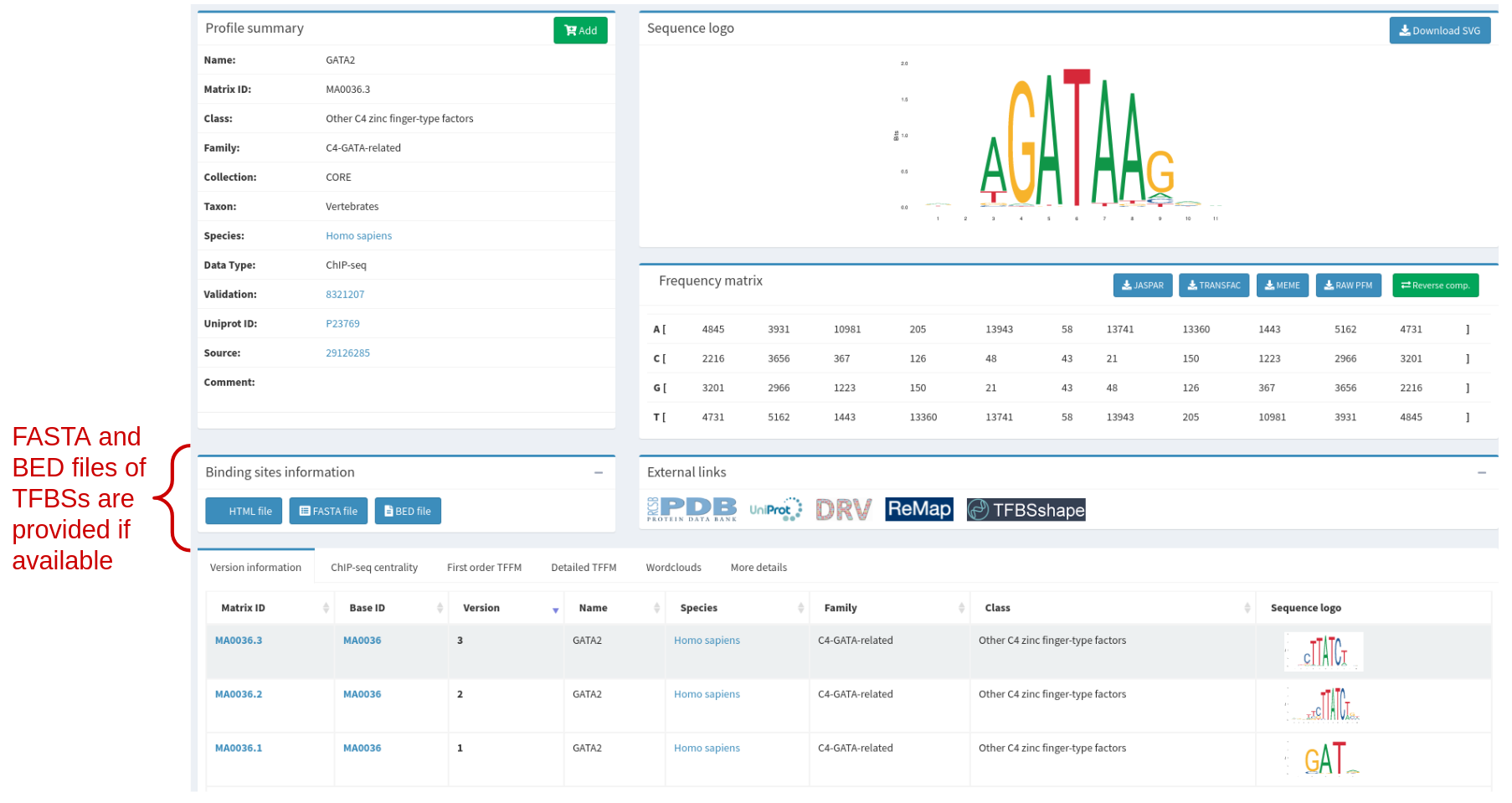
If a profile was obtained using our pipeline on ChIP-seq/exo data, the corresponding sequences can be downloaded in BED and FASTA formats for individual profiles. See an example figure below. All available sequences can also be downloaded in the Download Data section under “Other data”.
Why are certain sequences not downloadable from JASPAR CORE?
This is due to historical reasons. JASPAR CORE was originally built in order to create familial binding profiles for as many structural classes of transcription factors as possible. In some experimental literature, for example motifs generated from PBM and SELEX, only matrices and not sequences are available. For recent additions, for all the internally generated motifs (derived from ChIP-seq/-exo) it is a requirement to have the sequences (and the genomic coordinates) used to build the PFMs.
Why is my matrix study not included in JASPAR CORE?
There are two principal explanations. The most likely is that we were not aware of your work: please let us know! The other possible reason is that the publication did not live up to the demands of the curators. As we have human curation of all JASPAR CORE matrices, this is to some degree an arbitrary call – we are happy to discuss it with you.
Another possibility is that your motif is part of the Unvalidated collection where we store motifs without orthogonal literature support. In case your motifs of interest are part of the Unvalidated collection, please fill the fields provided at the ‘Community curation’ section at each motif page (e.g., a literature support for your motif) so our curation team can have a look and evaluate its further incorporation to the CORE collection.
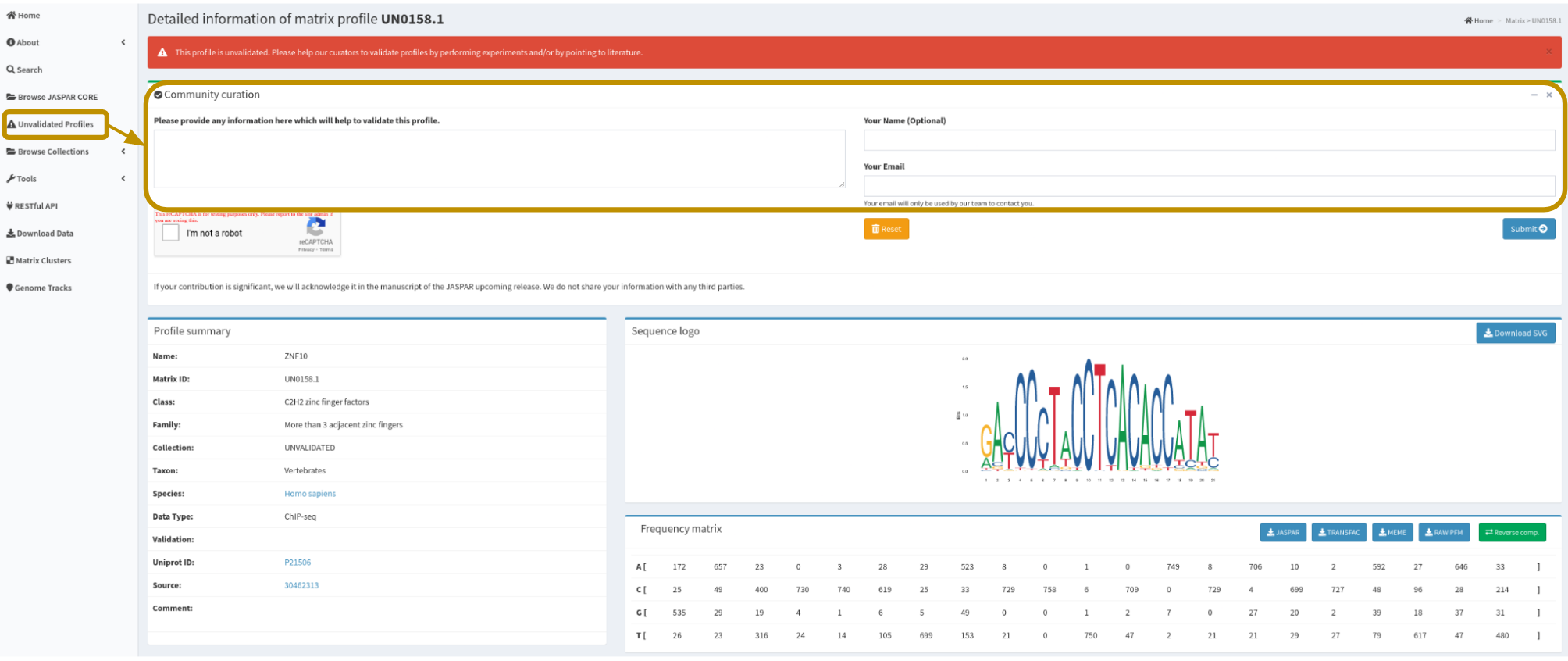
Each motif in the Unvalidated collection has a dedicated section where the user community may share information to add these motifs to the CORE collection in future JASPAR releases.
How to link web services to CPU-intensive services within JASPAR?
We appreciate that other services want to link to JASPAR. However, if your are using the CPU-intensive services (matrix comparison, randomization or clustering), please ask the maintainers (you can find contact information here) before you do this – otherwise your server might be rejected without warning. In that case, we strongly suggest setting up a local JASPAR database, as the database and resources are freely available.
Who is JASPAR anyhow?
JASPAR was originally the name of a master student project algorithm for comparing matrix profiles, an obscure tribute to an even more obscure dialog from the Black Adder episode “The Black Seal” between the Seven Most Evil Men in the Kingdom:
…and with all haste, we will meet at Old Jaspar’s tavern
How is old Jaspar these days?
Dead.
How?
I killed him.
[Loud cheer].
Did not find what you were looking for? You can also check or ask your question in the JASPAR Q&A Forum.
List of questions
- How do I cite JASPAR?
- What motif formats does JASPAR support?
- How are TFBSs for a profile in JASPAR computed?
- How should I interpret the TFBSs score?
- Where do I download TFBSs for a profile in JASPAR?
- Where do I download the sequences used to construct the JASPAR PFMs?
- Why are certain sequences not downloadable from JASPAR CORE?
- Why is my matrix study not included in JASPAR CORE?
- How to link web services to CPU-intensive services within JASPAR?
- Who is JASPAR anyhow?